

CYCELL uses the Cellular Potts Model (CPM). In a nutshell, the simulation runs in a ‘petri dish’ composed of a grid of squares.
We'll refer to each little square as a Node. Each Node has four neighbors in the North, East, West, and South directions.
A cell is nothing more or less than a collection of contiguous Nodes. Each Node is composed of four sides, called Membranes.
 For instance, in the diagram, all the yellow-colored Nodes
are considered a single cell. Ditto for the the blue-colored Nodes. (The white Nodes are empty and referred to as
the Medium.)
For instance, in the diagram, all the yellow-colored Nodes
are considered a single cell. Ditto for the the blue-colored Nodes. (The white Nodes are empty and referred to as
the Medium.)
The sin qua non of CPM is to swap Nodes in a particular manner such that cells change shape or 'move' around the petri dish in a realistic manner. During each iteration of the CPM simulation, every edge Node -- that is, a Node that is at the boundary of a cell -- is analyzed. Let's walk through this process with the edge Node marked with the red dot.
The program takes a look at the Node’s four neighbors, marked here as N, E, W, and S. It needs to decide whether to replace the marked edge Node with the contents of one of these neighbors. The decision to swap or not depends on many factors... but, the main two are as follows:
Each type of cell has an ideal Area, measured by the number of Nodes it contains. For instance, the yellow cell has an area of 9, and the blue cell an area of 3.
If a swap gets a cell's area closer to the ideal, the swap is more likely to occur.
Each cell has several adhesion factors. For instance a yellow-type cell might really really like to be next to a blue-type of cell. Or, maybe blue-type cells really want to have as many Nodes touching the Medium as possible. There are many possible combinations, and each possible swap will result in its own Adhesion score.
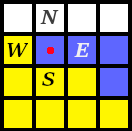 In short, with each iteration, each particular edge Node is assigned a score based
on its current Area, Adhesion (i.e., proximity to other cell-types), as well as several
other factors. The simulation tentatively swaps out the edge Node under consideration with each
of its four neighbors, scoring the resulting configurations using the same factors.
In short, with each iteration, each particular edge Node is assigned a score based
on its current Area, Adhesion (i.e., proximity to other cell-types), as well as several
other factors. The simulation tentatively swaps out the edge Node under consideration with each
of its four neighbors, scoring the resulting configurations using the same factors.
If a swap improves the overall score, it's kept. Otherwise, the status quo remains and
the program moves on to the next edge Node. In the diagram above, for instance,
perhaps the yellow cell was a bit
too large. And, perhaps the blue cell's Adhesion factor attracts it to the Medium. If so, it may
make sense to swap out the edge Node with the Node to the East, as shown to the right.
Occasionally, the system can get stuck in a local minimum. To deal with this, the program adds in a bit of randomness, or "Flux." With a high Flux, the program might keep a swap even it doesn't improve the score. This is necessary in order to get the system to work towards the ideal state; sometimes, you need to take a step or two back in order to move forward.
To learn more about CPM, we recommend you go straight to the source: Simulation of Biological Cell Sorting Using a Two-Dimensional Extended Potts Model. James A. Glazier, one of the article's authors, is the grand-daddy of CPM; his scholarly works are greatly appreciated by us at CYCELL, as are those of Marc Durand. Thanks guys!
After the program performs the CPM analysis and all the edge Nodes have been toyed with and re-adjusted, CYCELL then has each cell run the Rules. The Rules is the name for the little program each cell contains; you can write whatever Rules you wish. Each cell has the same Rules, just like real cells in an organism all have the same DNA. But, only certain parts of the Rules will be processed, depending on the cell's current state. The Rules control all kinds of behavior, such as migration, mitosis, and changing from one state to another.
After all the cells have processed their Rules, the CYCELL cycle is complete; the program performs a few housekeeping tasks behind the scenes, and then starts all over at the top.