

Referring to a program written within a program can easily lead to ambiguities.
As used herein, the term "The Rules" or "Rules" refers to the program you will be writing to control your cells, while the term "Program" refers to the CYCELL operating system.
Each Cell contains varying amounts of proteins and other substances that may affect the behavior of the Cell according to the Rules. For the CYCELL model, these proteins and other substances are treated as variables.
CYCELL uses four types of variables:
Some of these variables may be modified by the Rules. Others are read-only.
Prior to using the first three types of variables, they must be declared (i.e., given a name). The Internal variables are pre-named and ready for use, as described below.
A User variable (referenced herein as
A
Upon
A Gradient variable (referenced herein as
The
A cell may read only the
A Vector variable (referenced herein as
A
Internally, a
At any given time, a cell exists in a specific
A
Immediately following the declaration of a
A sentence is composed of conditions and commands. Alternatively, a sentence may have only commands.
Every sentence is formatted as follows:
Individual conditions and commands are separated by semicolons. The conditions, if any, are separated from the commands by a double colon (::).
At every cycle in the simulation, each cell will jump to that portion of the Rules that corresponds to its
Each sentence
will then be evaluated in the order it is written. Specifically, going
from left to right, the program will process each condition to determine if it evaluates
If any condition evaluates
If all the conditions evaluate IF statement:
IF ((condition_1)&&(condition_2)...&&(condition_n)){commands;}
Many of the conditions described below use logical comparisons between values. Take for instance
The logical comparison syntax is a little funky. But, it's simple to learn and very concise.
The bracket portion in the example above can be written 5 different ways. The possibilities are listed below, along with their logical meaning:
In order to express a > b (or a >= b) the keyword
In all the conditions below, unless otherwise noted, the bracketed portion may be swapped out for any of the above-listed possibilities.
All the conditions described below may be preceded by a !, which will cause a condition that would otherwise have evaluated
Many of the conditions and commands take parameters.
In the description of the syntax for each, the following convention will be used:
A condition triggered upon entry into the state.
It evaluates
In the following example:
when the cell first enters the Apple state, command_0 will be processed, and then command_1. The next time the cell is evaluated, the program will skip command_0 and proceed directly to command_1.
If the cell leaves the Apple state (through the
The condition is useful if you want something to occur every so often.
The first
(Likewise, command_1 will occur every cycle except, on average, every 23 cycles or so when it will be skipped. This is due to the inclusion of the !.)
The value of any literal_value, u_var, or g_var, may be compared to any other literal_value, u_var, or g_var.
In the example, Protein_a and Protein_b have been declared as
command_0 will be processed iff the value of Protein_a is greater than or equal to 2.34 AND less than 982.
command_1 will be processed iff the value of Protein_a equals the value of Protein_b.
command_2 will be processed iff the value of Protein_a >= 46 <= the value of Protein_b.
Note that the last sentence could also be written as follows:
A condition that counts adjacent cells.
The syntax is as follows:
Without the optional curly-bracket parameter,
With the optional curly-bracket parameter,
A condition that counts adjacent membranes.
The syntax is as follows:
Without the optional curly-bracket parameter,
With the optional curly-bracket parameter,

The term
A condition that evaluates adjacent membranes in terms of percents.
The syntax is as follows:
A condition based on where the cell thinks it is in the Field.
The syntax is as follows:
A condition based on where a cell thinks its edge Nodes are in the Field.
The syntax is as follows:
A command for setting the cell's color.
The syntax is as follows:
Parameters r, g, and b may be lliteral_values or u_vars. They correspond to the Red, Green, and Blue values of the cell's color.
All parameters will be clipped to values from 0 to 255.
A command for changing the cell's state.
The syntax is as follows:
This command sets the cell's
A command for setting a
The syntax is as follows:
This command will set the value of g_var in each of the cell's Nodes to 1.0.
After all the cells have run their Rules, the program diffuses the values of all the
A command for setting a
The syntax is as follows:
This command will set the value of v_var.
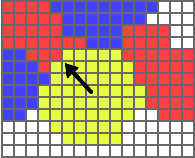 If the bracket contains a state_name, v_var will be set as the normalized sum of all vectors extending from the cell's center to each of the cell's Nodes
that touch a neighboring cell in
If the bracket contains a state_name, v_var will be set as the normalized sum of all vectors extending from the cell's center to each of the cell's Nodes
that touch a neighboring cell in
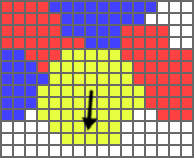 Similarly, if the bracket contains
Similarly, if the bracket contains
In the picture to the left, the black arrow represents v_var in such a scenario.
If the bracket contains u_var as the parameter, v_var will point towards the average position of all the cell's Nodes that touch adjacent cells containing u_var, with each such node weighted by the amount of u_var present.
If the bracket contains g_var as the parameter, the cell will consider the amount of g_var in the four Nodes to the North, East, West, and South of its center node and set v_var to point towards the average concentration of g_var in these Nodes.
Finally, the second form of the command takes two litaral values which represent the x and y components of the vector.
(This form is included primarily for debugging purposes and for use with the
The coordinate system used in CYCELL has a normal X-axis, with values increasing from left to right. But, the Y-axis is flipped; values increase from top to bottom. Thus, the vector (0.5, 0.5) points Southeast, the vector (0,1) points North, etc.
Regarding frames of reference, inputting (1,0) as a vector will result in a vector pointing East in the Petri Dish regardless of which way the cell thinks it is facing. And, if such a vector is used by
While it might be more realistic to define vectors in terms of a cell's frame of reference, we feel its easier for the user to keep the vectors pegged to the Petri Dish and allow the program to do all the rotations and whatnot behind the scenes.
A command for causing the cell to move.
The syntax is as follows:
The cell will move in the direction of v_var, the first bracket parameter.
The second bracket parameter indicates the 'strength' of migration. A negative number will induce movement in the direction of the vector (or rotated vector, as the case may be), while a positive number will induce movement in the opposite direction.
The third optional bracket parameter is an offset angle in degrees. If left blank, it will be 0. Otherwise, v_var will be rotated by the parameter's value.
There may be several
A command for causing the cell to split.
The syntax is as follows:
The first form will cause the mother cell to split along a random cleavage line. The daughter cell's state will be that of the mother.
The second form will cause the mother cell to split along a random cleavage line. The daughter cell's state will be state_name.
The third form will cause the mother cell to split along a cleavage line passing through the cell's center in the direction of v_var, offset by an angle given by the second parameter. The daughter cell's state will be that of the mother.
The final form will cause the mother cell to split along a cleavage line passing through the cell's center in the direction of v_var, offset by an angle given by the second parameter. The daughter cell's state will be state_name. The third parameter indicates which of the two resulting cells will be the daughter. Assuming the cleavage vector is pointing towards the top of the Petri Dish, "CW" would assign the portion to the right of the cleavage to the daughter, while "CCW" assigns the portion to the left to the daughter. (i.e. "clockwise" and "counter-clockwise.")
All

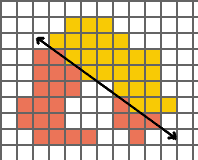

If a daugther would contain less than 5 Nodes, mitosis will not occur; but, it will be attempted on the next simulation cycle.
If the cell center lies outside the cell (a frequent occurence if the cell is stretched out) mitosis will not occur but will be attempted on the next simulation cycle.
Commands to directly modify
The syntax is as follows:
Parameter a can be any writeable variable. Parameter b can be any readable variable (other than a
The format is similar to many programming languages, wherein a+=b means a = a + b, and a-=b means a = a - b, etc.
Recall that no
Command to set a random float to a variable.
The syntax is as follows:
Parameter a can be any writeable variable. Parameters b and c can be any readable variable or a literal_value.
Parameter a will be set to a random float from the value of b up to c.
Command to kill the cell.
This command takes no parameters. Once processed, the program will exit the Rules.
Command to stop the Rules.
This command takes no parameters. Once processed, the program will exit the Rules.
There is an implicit
Command to make the cell sensitive to fields.
The syntax is as follows:
If the parameter is "1", the cell will sense fields. If "0", the cell will ignore fields.
Command to modify the Location component of the cell's Presumed Position.
The syntax is as follows:
a is a scaling factor and must be a literal_value.
b is a rotational offset value, measured in degrees, and may be a literal_value or u_var.
If left blank, a's default value is 1.0, and b's default value is 0.
The cell's current Presumed Position location will be modified by adding v_var, as modified by a and/or b, to it.
Simply put, if the v_var is a vector pointing due East, and a is equal to 1.0, the Cell's presumed location will move one unit to the East. Alternatively, one can think of the Field moving one unit to the West. The movement will be in the frame of reference of the Petri Dish and is irrespective of the Cell's presumed orientation; that is, in the preceding example, even if the cell's orientation was at 35 degree, the entire Field would nevertheless move 1 unit to the West.
Command to modify the Scale component of the cell's Presumed Position.
The syntax is as follows:
This command will create two sets of nodes.
The first consists of all the cell's edge nodes that are adjacent to
The second consists of all the cell's edge nodes that are adjacent to
The x and y scale components will then be adjusted to have the Field 'grow' in the direction of the first set, and 'shrink' in the direction of the second set. The literal_value represents the amount of the adjustment. The current

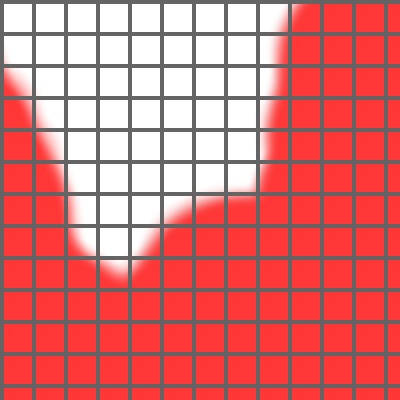

The literal_value parameter (we'll call it a) is related to the values of
where
Also keep in mind that the Program, at all times, will slowly adjust the x- and y-scale components of each cell's Presumed Position to have them gravitate towards the same value.
The first form dictates the degree to which cells of the same type adhere. The more negative the literal_value, the greater the adhesion.
The second form dictates the degree to which cells of the two different types adhere. The more negative the literal_value, the greater the adhesion.
The third form, using the keyword
The fourth form, using the keyword
The following values are defaults:
(The adhesion values of
The first form dictates the rate u_var will flow between cells of the same type. The rate is set by literal_value.
The second form dictates the rate u_var will flow between cells of type a and b. The rate is set by literal_value.
In both forms, the literal_value is the fraction of the u_var per shared membrane that will be transferred from one cell to the other every cycle.
For the following example,
A1 = A0 - (0.001 * MAB) + (0.001 * MAB × B0), where
A0 is the original level of protein in Apple,
A1 is the new level of protein in Apple after the iteration,
B0 is the original level of protein in Bear, and
MAB is the number of membranes shared between Apple and Bear.
The default rate for all junctions is 0. That is, unless set, no u_vars will be shared between cells.